Projects
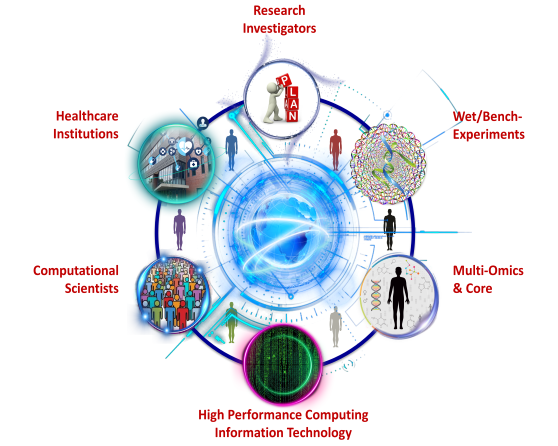
Precision Medicine
Precision medicine is driven by the paradigm shift of empowering clinicians to predict the most appropriate course of action for patients with complex diseases and improve routine medical and public health practice. We practice and promote integrating collective and individualized clinical data with patient specific multi-omics (e.g., genomics, transcriptomics, metabolomics) data to develop therapeutic strategies, and knowledgebase for predictive and personalized medicine in diverse populations.
Project details are availalbe at: Project Web Site
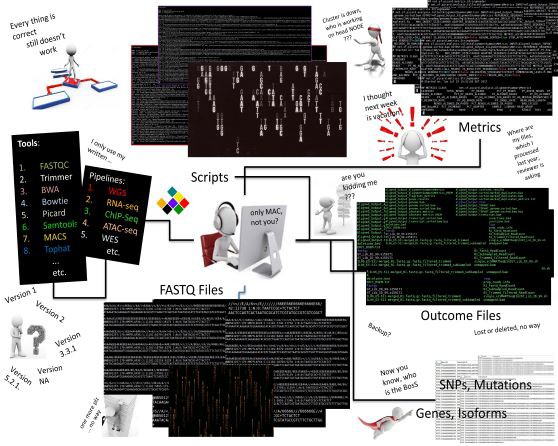
AI/ML & Bioinformatics
We are focused on dealing with unprecedented challenges in data science and provide better understanding of biology to revolutionize the field of medicine. Our ambitious research is underpinned with skills and resource development to build expertise in sequence-based genomic analysis, clinical variant interpretation, and evidence-based diagnostic and prediction model development and validation. We have developed and published many bioinformatics tools, genomics pipelines, gene-variant-disease annotation databases, and mobile health platforms to support clinical and multi-omics data analysis and dissemination.
AI/ML Applications:
- 3D IntelliGenes: AI/ML with multi-dimensional visualization
- IntelliGenes: AI/ML Desktop Software Application
- IntelliGenes: AI/ML Pipeline for biomarkers discovery and disease predictions
- Hygieia: AI/ML Pipeline
Bioinformatics Applications:
- VAREANT: Bioinformatics application for gene variant reduction and annotation
- GVViZ: Tool for gene disease annotation
- JWES: Pipeline for variant discovery
- PROMIS-APP-SUITE (PAS)
- I-ATAC
- Match & Scratch Barcodes
- GenomeVX
- Ant-APP-DB
- Dro-LIGHT-2
- Management, Analysis and Visualization of sequence data (MAV-seq)
Executable and source code of most of the applications are available at GitHub
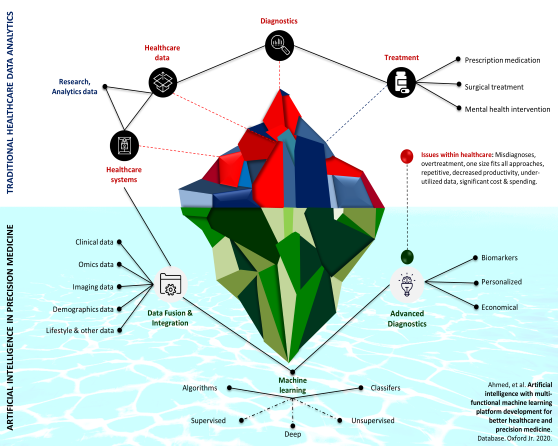
Healthcare Data Analysis
To implement effective precision medicine with enhanced ability to positively impact patient outcomes and provide real-time decision support, it is important to harness the power of electronic health records (EHR) by integrating disparate data sources and discovering patient-specific patterns of disease progression. We program intelligent, multifunctional and Health Insurance Portability and Accountability Act of 1996 (HIPAA)-compliant platforms for the efficient management and analysis of healthcare data. We have developed expertise in the electronic healthcare records (EHR) extraction, cleansing, restructure, encryption, aggregation and deidentification. Our implemented solutions have been succesfully tested using NextGen and EPIC health systems.
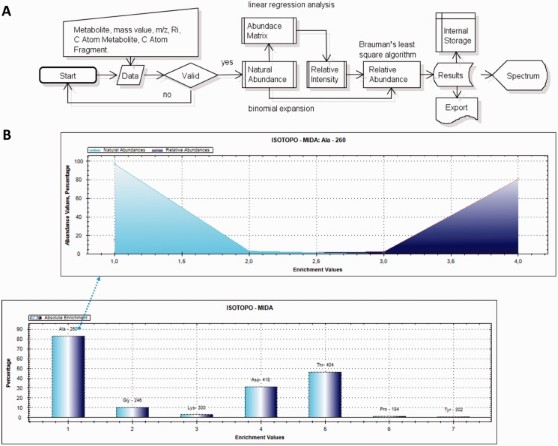
Metabolomics Data Analysis
Metabolites and their pathways are central for adaptation and survival. Metabolic modeling elucidates in silico all the possible flux pathways and predicts the actual fluxes under a given situation, further refinement of these models is possible by including experimental isotopologue data. We have established the key theoretical concepts and distinct analysis steps in the modeling process necessary to compare flux calculation and metabolite analysis. We are focused on designing systems for mass isotopomer distribution analysis and calculation of enrichments, which in-cluded bioinformatics applications (Isotopo and LS-MIDA) to analyze signal intensities from mass spec-tra of 13C-labelled metabolites, such as tert-butyldimethylsilyl-derivatives of amino acids. Furthermore, addressing a major challenge for mass spectrometric-based lipidomics, which is the need for rapid computational processing of the large amounts of data that are typically acquired when attempting to describe all the lipid species in a biological sample. We have developed a new bioinformatics application (Lipid-Pro) that generates discrete lipid profiles by interpreting datasets generated by liquid chromatography-tandem mass spectrometry using the advanced data-independent acquisition mode MSE.
- Lipid-Pro: lipid identification solution for untargeted lipidomics.
- Isotopo: mass isotopomer data analysis and management.
- LS-MIDA: mass isotopomer distribution analysis in metabolic modelling.
- Quantitative metabolic flux analysis and modeling.
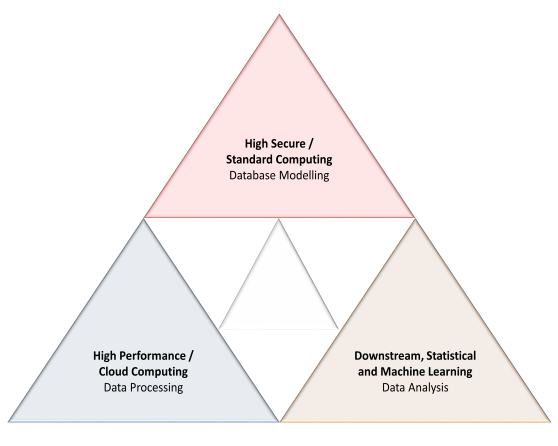
Computing Infrastructures
To support high-volume and heterogenous data management, sharing, processing, and intelligent and integrated analysis, we proposed, modelled, and successfully implemented big data infrastructure. Overall infrastructure is based on five independent and distributed data servers:
- High Performance Computing (HPC) based data server for efficient and timely big data processing;
- Relational database management server (RDBMS) for maintaining processed data and supporting downstream data analysis, visualization and sharing;
- Virtual data server to support integration between HPC and RDBMS servers for efficient data Extraction, Transfer and Load (ETL);
- Web and database servers to support sharing of information and knowledge at global level via smart phone or iOS applications, and websites;
- Data server to support sharing of open-source code and script to help researchers with in Rutgers and even research communities at national and international lever.